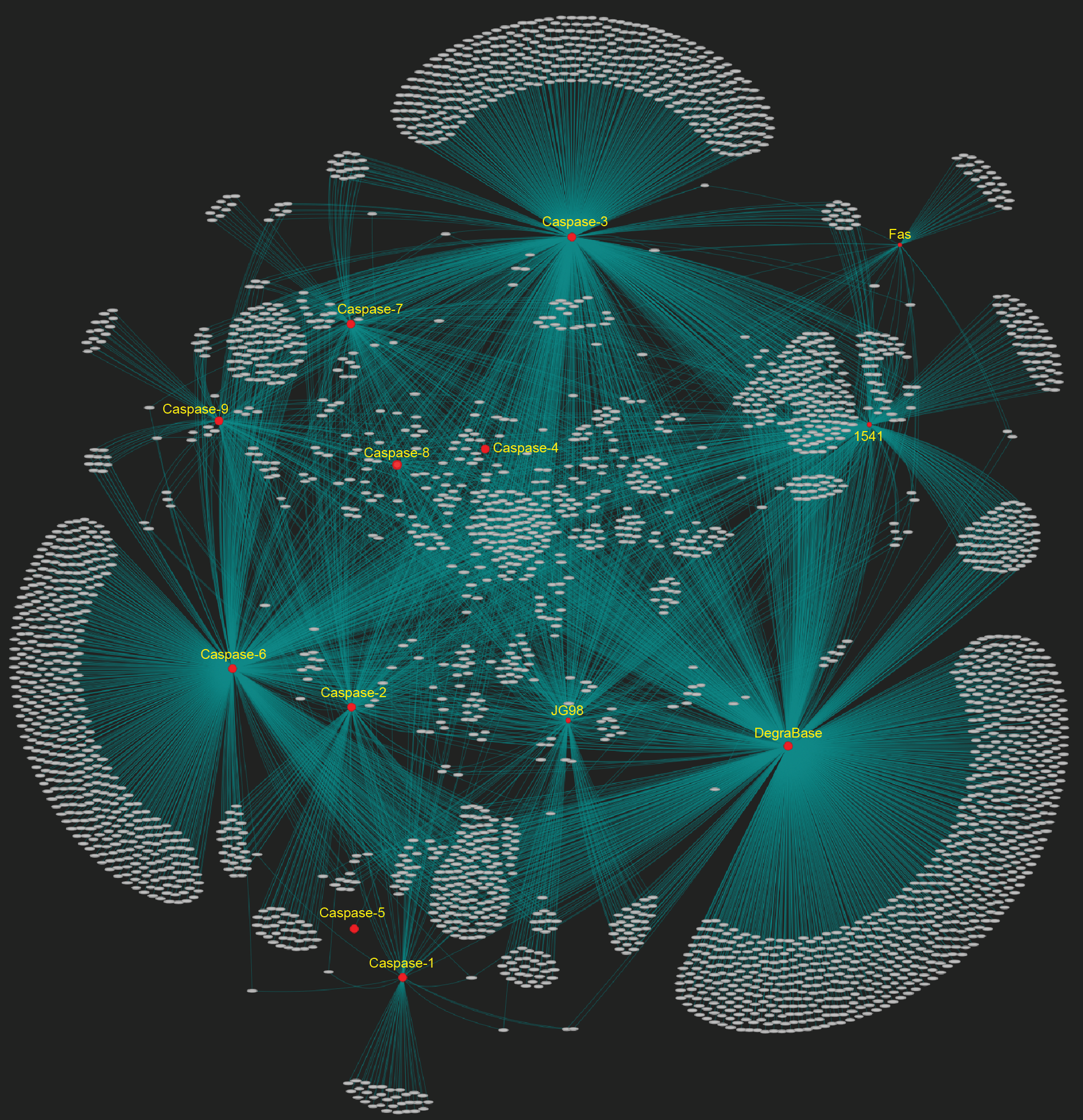
CaspSites is a collection of experimentally identified human caspase substrates observed in apoptotic cells and lysates. Stored data was collected from a number of published studies using N-terminal labeling techniques to identify proteolytic events in human cell lines (see below for data set breakdown, their overlap, and references).
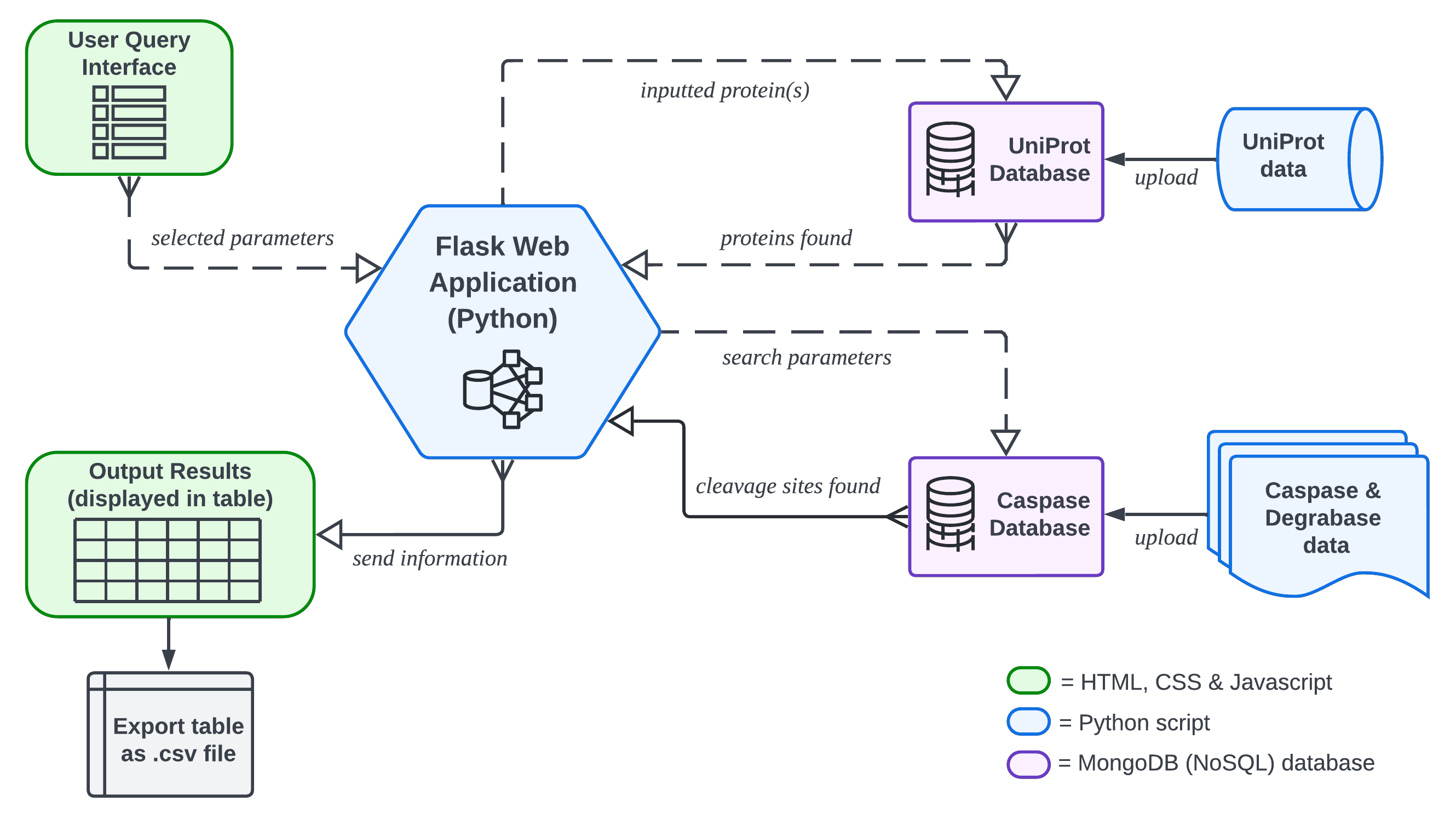
Users can perform queries on CaspSites content by entering lists of proteins and selecting P4-P4' site residues to search custom-selected datasets. Common substrates between multiple caspase data sets can be found by pressing the logical operator toggle found on the search page. A workflow diagram of the operations performed and layout of CaspSites can be seen on the right.
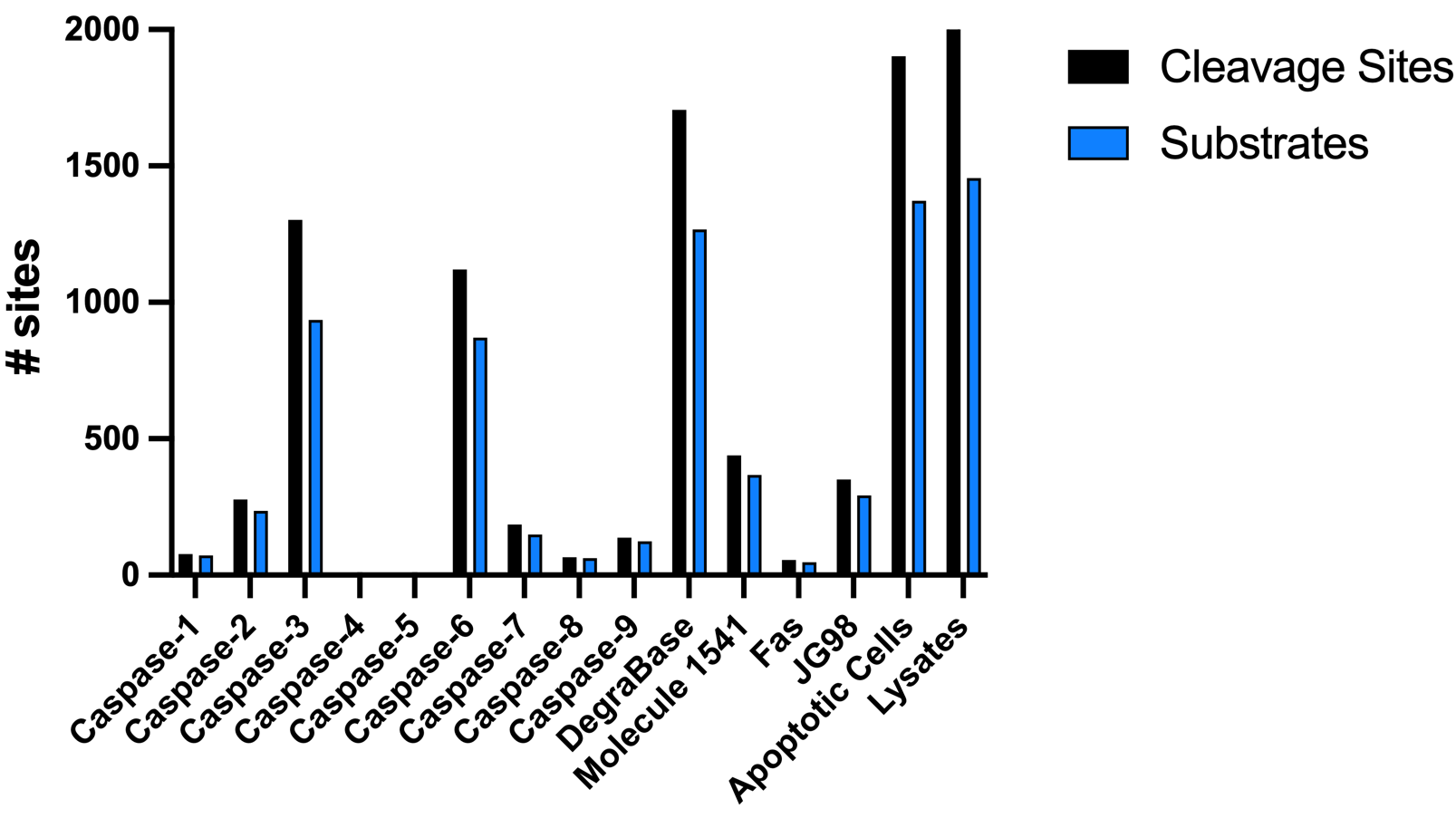
| Data Set | # Cleavage Sites | # Substrates |
|---|---|---|
| Caspase-1 | 78 | 73 |
| Caspase-2 | 300 | 253 |
| Caspase-3 | 1197 | 957 |
| Caspase-4 | 2 | 2 |
| Caspase-5 | 1 | 1 |
| Caspase-6 | 1120 | 871 |
| Caspase-7 | 211 | 190 |
| Caspase-8 | 65 | 62 |
| Caspase-9 | 137 | 124 |
| DegraBase | 1706 | 1268 |
| Molecule 1541 | 439 | 368 |
| Fas | 55 | 47 |
| JG98 | 350 | 293 |
| Apoptotic Cells | 1902 | 1373 |
| Lysates (Caspase 1-9) | 2028 | 1456 |
If you would like to directly access data stored in CaspSites (read-only),
click here for more information about the programming languages and APIs supported by MongoDB.
- Please use the following access information (case-sensitive) - contact us to learn more about database structure:
| Username | Password | Connection String | Database Name | Collection Name |
|---|---|---|---|---|
| CaspSitesGuest | caspase | mongodb+srv://<username>:<password>@julienlab.fibou.mongodb.net/?retryWrites=true&w=majority | CaspSitesGuest | CaspSitesData |

 Created by Henry Wang @ Julien Lab, Department of Biochemistry
Created by Henry Wang @ Julien Lab, Department of Biochemistry
