Exclude the following datasets from searches: (hold control to select multiple)
Choices above will be used to exclude dataset substrates (P1 = D) from searches. For example, if the caspase-1 dataset is chosen for the search and caspase-2/-3 substrates are excluded,
the output will only show substrates unique to caspase-1 not seen in caspase-2/-3 experiments.
Cleavage Site Information:
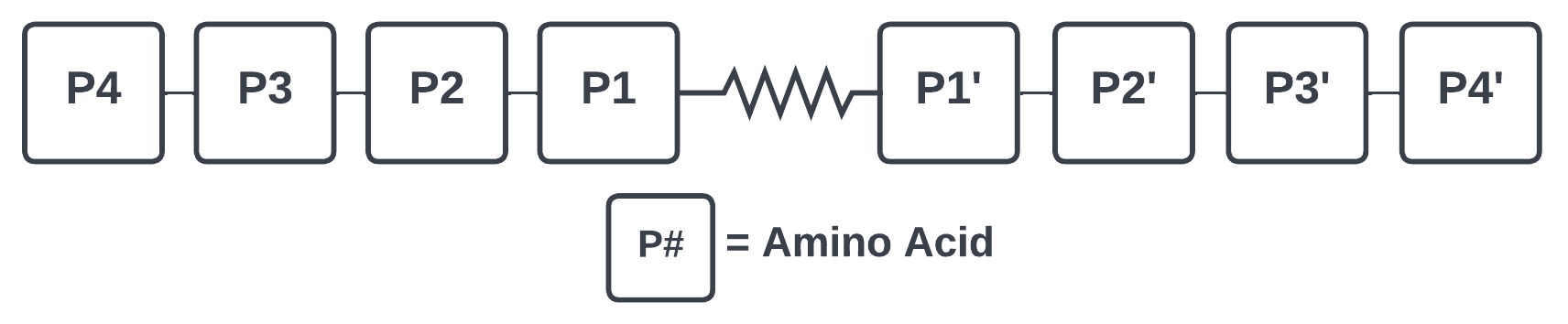
Protease subsite nomenclature is reported as the P4-P4' site
(Schechter & Berger, 1967).
The four amino acids prior to cleavage of the peptide bond (N-terminal) are designated as the P4, P3, P2, and P1 residues.
Similarly, the four residues after the cleaved bond (C-terminal) are designated as P1', P2', P3' and P4'.
- Caspases preferentially cleave substrates after aspartic acid residues (P1 = D)
 Created by Henry Wang @ Julien Lab, Department of Biochemistry
Created by Henry Wang @ Julien Lab, Department of Biochemistry
